How to SNAPP a bacterial cell
Computer simulations are helping develop a new generation of bacteria-busting molecules and demonstrating the deadly power that could help solve the antimicrobial resistance crisis
Published 18 June 2025
Imagine a bacterial cell – one of the multi-drug-resistant varieties that keep infectious disease experts up at night – blown apart like a microscopic firecracker.
That’s exactly what scientists are seeing when a new kind of molecule called SNAPP meets bacteria.
It’s not science fiction – it’s molecular dynamics in action.
Our team is using molecular dynamics computer simulations to understand how SNAPPs – Structurally Nanoengineered Antimicrobial Peptide Polymers – destroy drug-resistant bacteria.
We’re hoping to change the game in the battle against the escalating global health crisis of antimicrobial resistance.
A jellyfish that kills bacteria
SNAPPs resemble tiny jellyfish – soft, spherical bodies with wavy amino acid ‘arms’ that reach out and interact with the membranes that surround bacterial cells.
And those arms are deadly.
Even at low concentrations, SNAPPs induce rupture, collapse and disintegration of bacterial cell membranes. Rather than simply attacking the bacteria, SNAPPs destabilise the fundamental structural integrity of the membrane.
This eliminates any chance of the bacteria developing resistance against SNAPPS.
Our collaborators, Professors Greg Qiao and Neil O’Brien-Simpson pioneered the synthesis of SNAPPs, first demonstrating their antimicrobial activity in the laboratory.
But not all SNAPPs behave the same way. Small changes in the sequence, type and number of their amino acid arms can change how effectively they kill bacteria – and whether they might also harm human cells.

And there are lots of possible SNAPPs.
There are 20 types of amino acids to choose from and there are hundreds spread across the eight arms of each SNAPP molecule. That means countless possible combinations.
It’s a molecular recipe with more variations than any kitchen cookbook.
Building a smarter way to design molecules
That’s where our team comes in.
At the Soft Matter Informatics group, we combine computational materials science with machine learning and eXplainable Artificial Intelligence (XAI) to tackle complex design challenges like SNAPPs.
We’ve built computer simulation models that let us see what no experiment can: the moment-by-moment interaction between a SNAPP molecule and a bacterial cell membrane at the nanoscale (billionths of a metre).

PhD student Amal Jayawardena has spent the past couple of years building detailed models that reveal how SNAPP arms sink into bacterial membranes, fold around them and essentially tear them apart from the inside.
It’s stunning to watch and it illustrates why even a few SNAPP molecules can be lethal to bacteria.
Just as important, we can see how these molecules don’t latch onto mammalian cells the same way. That’s because bacterial membranes carry a distinct surface charge – a feature our cells lack.
That’s part of what makes some new variants of SNAPPs so promising as targeted antimicrobials.
Enter Bandicoot: AI meets molecular design
Of course, simulating one SNAPP at a time won’t get us far.
There are too many possible combinations. We need a system that knows where to look and what to look for.
So, we’re building it.

Sciences & Technology
The vaccine improving the health of Australia's chickens
We call it Bandicoot. We've named it after the clever little marsupials that poke around in the soil, sniffing out what’s hidden beneath the surface.
That’s what Bandicoot does: it explores complex data, identifies where our knowledge is thin and directs simulations and experiments to the most promising areas.
Postdoctoral researcher Dr Nick Robe built a software platform to generate simulations for industrial polymers. We’ve adapted it to create thousands of different SNAPP variants.
And PhD student Jalal Abdolahi has developed ‘intelligent search’ algorithms – tools that help Bandicoot decide which SNAPPs are worth simulating next.
If some variations aren’t changing outcomes, we don’t waste time on them. Instead, we focus where the science is hottest.
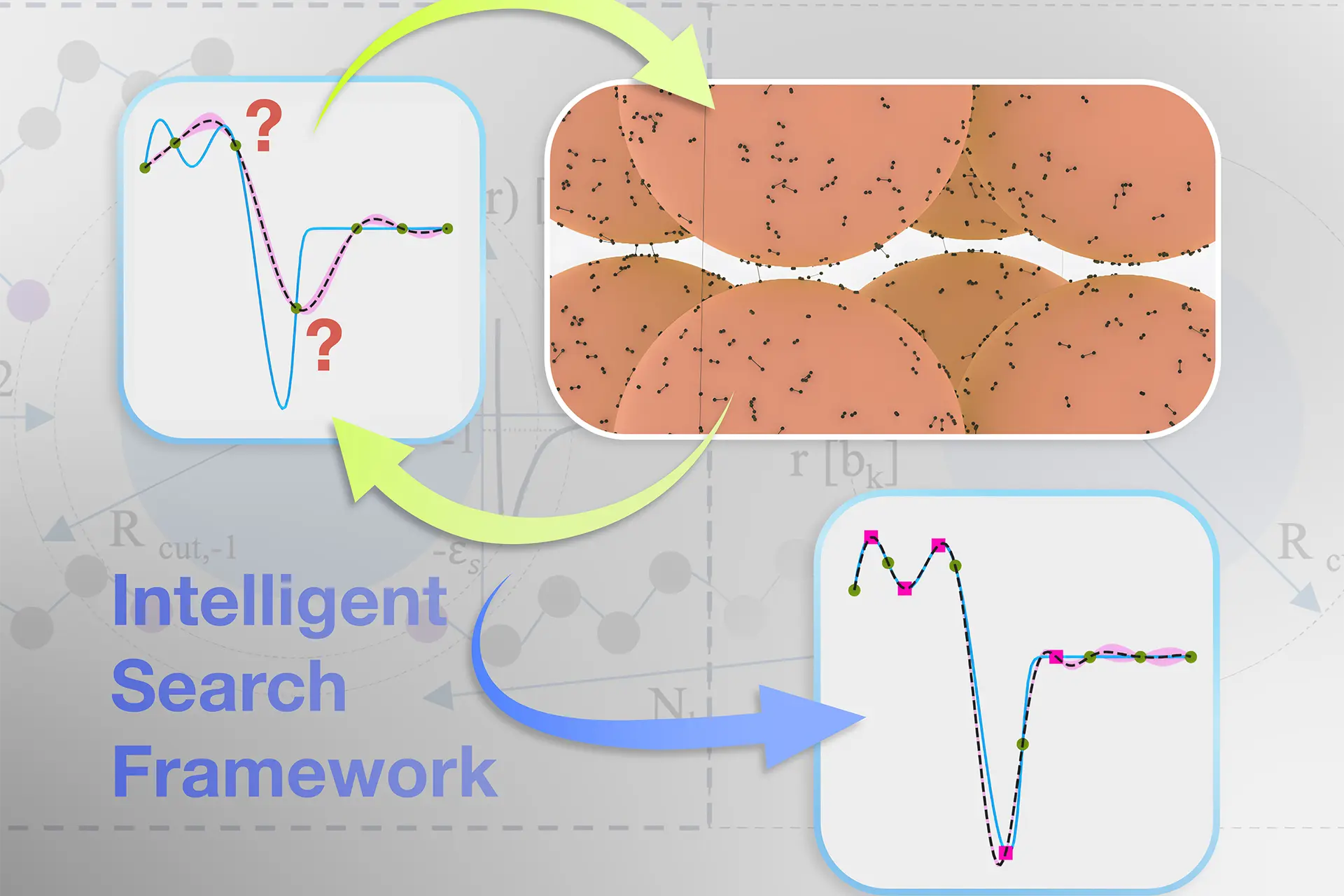
More than a ‘black box’
But we don’t just want Bandicoot to tell us what works. We want it to tell us why.
That’s where explainable AI comes in.
Instead of a black box that spits out answers, Bandicoot gives us insights – explaining which parts of a SNAPP molecule make it effective and how those features interact.
That kind of interpretability is critical. It helps scientists like us understand the fundamental rules behind molecular behaviour.
It also has the potential to help product developers create safer, more reliable treatments.
These explanations don’t just satisfy curiosity – they also help speed up the discovery process.
Sciences & Technology
We now know what causes the Buruli ulcer, so what are we doing about it?
Turning the tide
Antibiotic resistance is a global health crisis. But with tools like SNAPPs and Bandicoot, we’re shifting from reaction to prediction – from fighting fires to designing fireproof systems.
For the first time in decades, the upper hand may be within reach.
Smart molecules made by smarter systems, with the potential to outsmart superbugs at their own game.
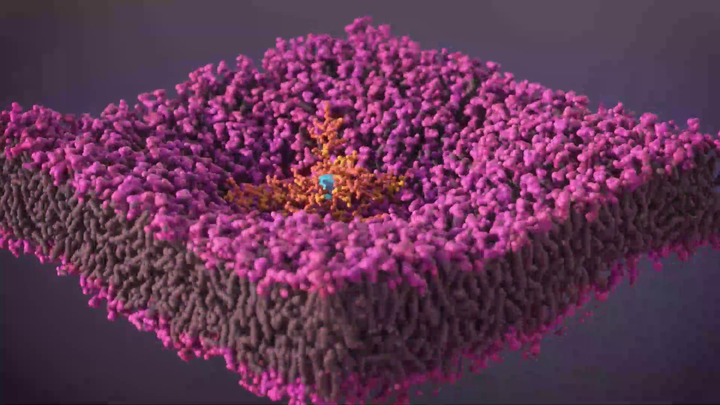
The banner video shows the step-by-step process of membrane disruption due to SNAPP insertion into the membrane bilayer, leading to bacterial cell death. Visualisation by Luke Richardson, based on MD simulation data by Amal Jayawardena.
The parallax video demonstrates how lipidation (blue colour) significantly enhances the antimicrobial activity of SNAPP molecules against bacterial membranes through improved membrane interactions, deeper insertion, stabilised binding, and more effective membrane disruption. Video: Amal Jayawardena



