
Health & Medicine
Using chemotherapy to fight malaria

Over the last 50 years, malaria parasites have developed resistance to seven drugs, but a new way to identify future antimalarials holds promise
Published 3 June 2022
So far, the malaria parasite has had an answer to every method of control we have thrown at it.
Despite significant advances in the fight against it - including a vaccine roll-out that held promise, but only shows about 30 per cent effectiveness against severe disease - and dozens of drugs – the parasite always makes a comeback.
Currently, the recommended treatment for malaria infection is an artemisinin combination therapy, a mix of fast and slower-acting drugs designed to treat malaria infection and prevent transmission.

However, combination therapies fail to cure infections in more than 50 per cent of patients in some Southeast Asian regions. Furthermore, resistance to artemisinins has also now been detected in Africa, where most of the 600,000 deaths a year caused by malaria occur.
To help save lives and improve quality of life, breakthrough drugs that hit new targets and use new mechanisms of action are desperately needed.

Health & Medicine
Using chemotherapy to fight malaria
Professor Leann Tilley, from the Department of Biochemistry and Pharmacology, is part of an international research team working to address global concerns that current antimalarial treatments are rapidly losing their effectiveness.
Professor Tilley and colleagues have published a world-first discovery in the journal Science showing that a previously overlooked class of chemicals – known as nucleoside sulfamates or “nukes” – can cause malaria parasite enzymes involved in protein synthesis to self-destruct.
These new targets are very effective because protein synthesis enzymes play critical roles in the maintenance and growth of cells.
Of particular importance, inhibitors of this pathway are expected to be active against all stages of the malaria parasite – effective for both treatment and preventing transmission to new victims.
Specifically, the nucleoside sulfamates were found to hijack the parasite’s own cell machinery, inducing enzymes involved in protein production to create their own inhibitors – thereby halting processes that are essential for the parasite’s survival.
This mechanism had not been previously reported for protein synthesis enzymes.
“In short, we discovered a new avenue for targeting pathogens – getting them to be the instrument of their own demise,” says Professor Tilley. “The prevention of transmission is very exciting as this will help slow the development of resistance.”
Working with the peak body for antimalarial drug development, Medicines for Malaria Venture and Takeda Pharmaceuticals, as well as research labs from across five continents, the large international team began their research with compounds that Takeda was investigating to treat cancer.

Health & Medicine
Killing the malaria parasite by blocking its recycling system
The team identified a series of compounds that affect the malaria parasite but not human cells – but the mechanism of toxicity was not understood.
“We were fortunate to have access to the suite of biochemical and structural biology platforms at the University of Melbourne’s Bio21 Institute of Molecular Science and Biotechnology, as well as The Australian Synchrotron,” says Professor Michael Parker, Bio21 director.
“That allowed us to mount a multi-pronged investigation of the mechanism of action.”
Further work by Dr Stanley Xie and Dr Elyse Dunn, from the Department of Biochemistry and Pharmacology, working closely with colleagues from Takeda Pharmaceuticals, allowed the team to discover that nucleoside sulfamates hijack protein synthesis enzymes to form covalent inhibitor-amino acid conjugates – a bit like super-gluing a key into a lock so that the lock no longer functions.
“Excitingly, we discovered a particular compound, ML901, in the Takeda compound library that targets a single plasmodium enzyme and was non-toxic to mammalian cells,” says Professor Tilley.
Colleagues from the Department of Biochemistry and Pharmacology, Dr Riley Metcalf, Dr Craig Morton and Associate Professor Mike Griffin solved the protein’s structure.
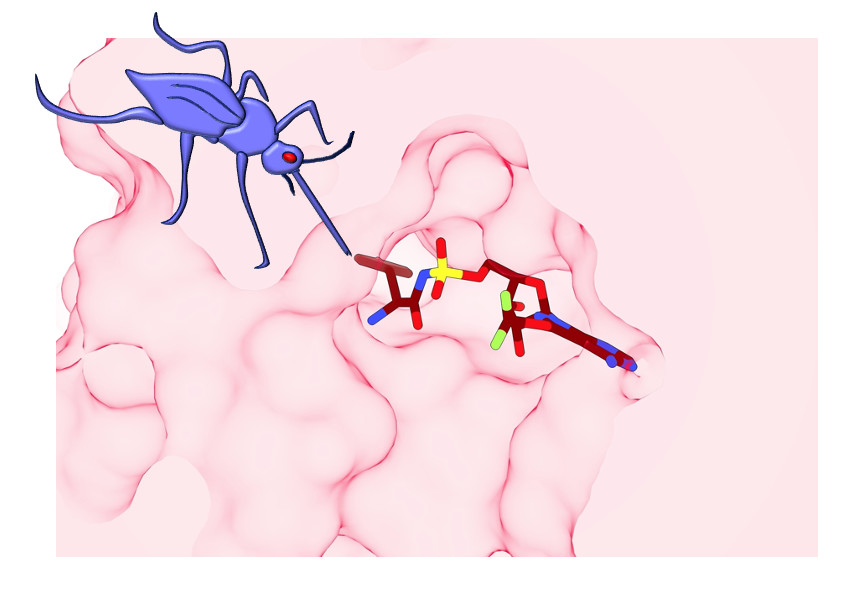
“We discovered a flap of protein that sits over the site where the inhibitor-conjugate binds,” explains Associate Professor Griffin. “The flap appears to hold the working enzyme in an intermediate state that is poised for attack by ML901.

Health & Medicine
Immunity to COVID-19: Lessons from malaria
“The human enzyme has a much more open active site, which means it is less susceptible to reaction hijacking by ML901. Our 3D views of the active site were very important to understand why ML901 is so potent and selective.”
The next phase was to test ML901 in a suite of malaria assays provided by the Medicines for Malaria Venture. These assays are designed to ensure that drug candidates meet the criteria for further development.
The team showed that ML901 is active against all stages and strains of the malaria parasite tested. Importantly, ML901 exhibits rapid and prolonged activity that gives potent parasite killing in an animal model of human malaria meeting the criteria for fast and effective treatment of malaria patients.
“The team is now ready to pursue the development of ML901 as a new antimalarial drug candidate,” says Professor Tilley.
With at least 200 million new malaria infections diagnosed each year, it’s hoped this class of nucleoside sulfamates will have similar success to other nucleoside sulfamates that target a different class of enzymes called ubiquitin activating (E1) enzymes.
These compounds have been exploited by Takeda to develop multiple new anti-cancer clinical candidates.

“We believe our work here is just the beginning,” says Dr Larry Dick, an Honorary Fellow in the Department of Biochemistry and Pharmacology, and co-lead author.
“This opens up several important new drug discovery avenues to help address the deadly impact of malaria and other infectious diseases - particularly in developing nations. It could also be used to target other diseases like cancer, neurodegenerative disease, metabolic syndromes including, diabetes and autoimmune disorders.”
According to Professor Tilley, the next step for the team is to tweak the chemical structure to improve the drug-like properties to optimise the absorption and distribution of the compound in the body.
“A particularly exciting part of the work is the ability to work with a large, talented and highly collaborative team, bringing together scientists from academia, the pharmaceutical industry and the not-for-profit sector.”
This research is funded by the Global Health Innovative Technology (GHIT) fund. The program has received approval from the Medicine for Malaria Venture to enter lead optimisation phase and the team will now seek funding from GHIT for the next development phase.
Banner: Getty Images